The Art of Transcript Editing: Exploring the Fascinating World of RNA Splicing
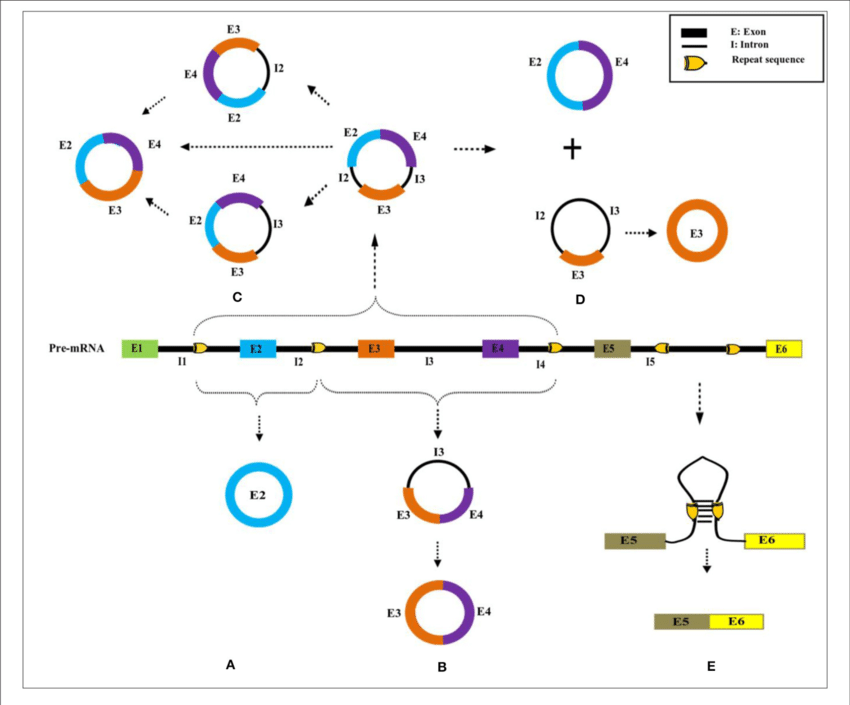
RNA splicing is a fundamental and intricate process in the realm of molecular biology, essential for the accurate expression of genes. This sophisticated mechanism involves the precise removal of non-coding regions, known as introns, from precursor messenger RNA (pre-mRNA) and the subsequent joining of coding sequences, or exons, to form mature mRNA molecules. This mature mRNA is then translated into proteins, the workhorses of the cell.
The Complex Process of RNA Splicing
The process of RNA splicing is carried out by a dynamic complex called the spliceosome, which consists of small nuclear RNAs (snRNAs) and associated proteins. The spliceosome orchestrates a series of highly regulated steps to ensure the correct excision of introns and the ligation of exons:
- Recognition and Assembly: The spliceosome assembles on the pre-mRNA at specific sequences near the intron-exon boundaries, known as splice sites. These sites are marked by conserved nucleotide sequences that signal the start and end of introns.
- Introns Removal: The spliceosome catalyzes two transesterification reactions. In the first step, the 5' splice site is cleaved, and a lariat structure is formed when the intron loops back and binds to a branch point within itself. In the second step, the 3' splice site is cleaved, and the exons on either side of the intron are joined together.
- Exon Ligation: The result is a continuous coding sequence free of introns, which can then be exported from the nucleus to the cytoplasm for translation into a protein.
Importance of Correct Splicing for Gene Expression
Correct splicing is crucial for the accurate expression of genes. Each mRNA transcript must be spliced correctly to produce a functional protein. The diversity of proteins that a single gene can produce is greatly enhanced by a phenomenon known as alternative splicing. This process allows different combinations of exons to be joined together, generating multiple mRNA variants from a single pre-mRNA transcript. Consequently, a single gene can give rise to different protein isoforms, each with potentially distinct functions and regulatory properties.
Consequences of RNA Splicing Errors
The precision of RNA splicing is critical, and errors in this process can lead to severe consequences, including various diseases. RNA splicing errors can result from mutations in the splice sites, errors in the spliceosome components, or defects in the regulatory mechanisms controlling splicing. These errors can cause the inclusion of introns or the exclusion of exons in the mature mRNA, leading to the production of malfunctioning or deleterious proteins.
RNA Splicing and Disease
One of the most prominent examples of diseases caused by splicing errors is cancer. Mutations that affect splicing can lead to the aberrant expression of oncogenes or tumor suppressor genes. For instance, mutations in the splicing factor SF3B1 have been implicated in various cancers, including chronic lymphocytic leukemia and myelodysplastic syndromes. These mutations often result in the mis-splicing of critical genes involved in cell cycle regulation and apoptosis, contributing to uncontrolled cell proliferation and tumor development.
Beyond cancer, splicing errors are also implicated in numerous genetic disorders. Spinal muscular atrophy (SMA) is a notable example, where a mutation in the SMN1 gene affects the splicing of its mRNA, leading to the loss of motor neurons and severe muscular atrophy.
Process of RNA Splicing
1.Recognition of Splice Sites:
- The spliceosome recognizes specific nucleotide sequences at the intron-exon boundaries, known as splice sites. The 5' splice site marks the beginning of an intron, while the 3' splice site marks the end. Additionally, a branch point sequence within the intron is crucial for splicing.
2.Assembly of the Spliceosome:
- The spliceosome assembles on the pre-mRNA in a stepwise manner. U1 snRNP binds to the 5' splice site, and U2 snRNP binds to the branch point sequence. This assembly is followed by the recruitment of the U4/U6.U5 tri-snRNP complex, forming the mature spliceosome.
3.Catalytic Steps of Splicing:
- The spliceosome undergoes conformational changes to bring the splice sites into close proximity. The 2' hydroxyl group of the branch point adenosine attacks the 5' splice site, forming a lariat structure. Subsequently, the 3' hydroxyl group of the upstream exon attacks the 3' splice site, resulting in the ligation of exons and the release of the intron lariat.
4.Release of Spliced mRNA:
- After the intron is removed, the exons are joined together, and the mature mRNA is released. The spliceosome disassembles, and the snRNPs are recycled for further splicing events.
Conclusion
RNA splicing is a meticulously regulated process essential for the correct expression of genes and the proper functioning of cells. The removal of introns and the precise joining of exons are critical for generating functional proteins. However, errors in RNA splicing can have devastating effects, leading to a range of diseases, including cancer. Understanding the complexities of RNA splicing and its regulation continues to be a major focus of molecular biology, with significant implications for the development of therapeutic strategies aimed at correcting splicing defects.